|
ORIGINAL ARTICLE |
https://doi.org/10.5005/jp-journals-10082-03191
|
Comparison of Extraction Free Dry Swab with Standard Extraction-based RT-PCR Method for Diagnosis of SARS-CoV-2
1Infectious Diseases, Thyrocare Technologies Limited, Navi Mumbai, Maharashtra, India
2,5,7Department of Community Medicine, Hinduhridaysamrat Balasaheb Thackarey Medical College and Dr. Rustom Narsi Cooper Municipal General Hospital, Mumbai, Maharashtra, India
3Thyrocare Technologies Limited, Navi Mumbai, Maharashtra, India
4,6Molecular Biology, Thyrocare Technologies Limited, Navi Mumbai, Maharashtra, India
Corresponding Author: Smita Santosh Chavhan, Department of Community Medicine, Hinduhridaysamrat Balasaheb Thackarey Medical College and Dr. Rustom Narsi Cooper Municipal General Hospital, Mumbai, Maharashtra, India, Phone: +91 9320509320, e-mail: drsmita1409@gmail.com
How to cite this article: Nikam C, Chavhan SS, Sengupta C, Ganesan P, Adsul B, Narkhede C, et al. Comparison of Extraction Free Dry Swab with Standard Extraction-based RT-PCR Method for Diagnosis of SARS-CoV-2. J Basic Clin Appl Health Sci 2023;6(2):29–32.
Source of support: Thyrocare Technologies Limited
Conflict of interest: None
Received on: 19 January 2023; Accepted on: 05 March 2023; Published on: 13 April 2023
ABSTRACT
Background: Coronavirus disease-19 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which had become a global pandemic leading to a great loss of lives and severe social and economic disruption. Accurate identification of the infected individuals is needed to control and reduce the spread of infection. Recently, an advisory has been issued by the Indian Council of Medical Research (ICMR) for using extraction free dry swab (EFDS) protocol to detect SARS-CoV-2 (COVID-19) with the aid of real-time reverse-transcription polymerase chain reaction (PCR).
Materials and methods: During the study, we have done a comparison of two methods on 500 clinical samples. Results obtained with EFDS were compared using the extraction-dependent viral transport medium (VTM) as a comparative method for the diagnosis of SARS-CoV-2.
Results: The overall performance of the EFDS method while compared with the comparative method showed 59 discordant results with a sensitivity of 85.65% and specificity of 98.47% with an accuracy of 92.48%.
Conclusion: This study shows that the EFDS shows comparable results with extraction-dependent VTM as a comparative method for the detection of SARS-CoV-2, dry swab method reduces and simplifies the workflow, reduces turnaround time, and is cost-effective.
Keywords: COVID-19, Dry swab, Extraction free, Pandemic, Reverse-transcription polymerase chain reaction, Severe acute respiratory syndrome coronavirus 2.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative pathogen of the (COVID-19). In order to reduce the magnitude of the disease, measures like promoting quick diagnosis, early initiation of treatment, strengthening of quarantine facilities, and improvement in isolation have been advocated. The implementation of the above prevention and control measures can significantly aid in ensuring optimal containment of the viral infection.1 False- negative/positive polymerase chain reaction (PCR) results must be ruled out by every laboratory and specific and accurate result must be provided so that the potential risk of transmission to household contact and community can be averted. This calls for the need that we must ensure appropriate sampling of specimens to facilitate the diagnosis of the novel viral infection.2
Collecting the appropriate specimen at the right time and transporting it to the laboratory under proper conditions are critical pre-analytical components of the testing process. The type of specimen collected will depend to some extent on the authorized SARS-CoV-2 viral test used. Such specimens may include healthcare personnel (HCP)-collected nasopharyngeal (NP), and oropharyngeal (OP) swabs.3 It should be made of synthetic fibers and have plastic or wire shafts. Besides this, there are new advisories by Indian Council of Medical Research (ICMR) suggestive of extraction-free protocol excluding VTM; dry swabs (DS).4 Considering the fact that the number of reported cases and the workload in the laboratory is extremely high, there have been issues with maintaining the supply chain and management, especially of the laboratory consumables. Moreover, we cannot rule out the shortage reported in the specimen collection sets and RT-PCR reagents.5 It is also important to note that a large number of national laboratories have been established that have been sharing the burden of laboratory consumables and other logistics.6,7 Many kit producers eventually made their way into the expanding market with readily available kits. Due to the biomedical industry’s quick response and the expanding number of RT-PCR test kits available, there has been a gradual move towards utilizing conventional equipment to conduct RNA extraction.
Thus, the present study has been planned with the intention to assess the effectiveness of an extraction free dry swab (EFDS) protocol with extraction dependent viral transport medium (EDVTM) for the detection of the causative virus by RT-PCR; which has recently been used often to collect samples for the purpose of SARS-CoV-2 molecular detection. The effectiveness of the transport medium and dry swab was assessed by COVIPATH RT-PCR kit (Thermo Fisher Scientific). In this study, we have compared two methods on 500 clinical samples with EFDS and EDVTM protocol; as the reference method for diagnosis of SARS CoV-2 by RT-PCR.
MATERIALS AND METHODS
Settings
In our study, the thyrocare service providers (TSPs), at Seven Hills Hospital, Mumbai, were involved in the process of collection of samples. The central processing laboratory (CPL) of thyrocare received samples that were transported at the desired temperature with mandate work order entries. At CPL, Thyrocare Technologies Limited, Navi Mumbai, India, sample storing, RNA extraction, PCR setup, and data analysis were performed.
Ethics
This study is approved by the Seven Hills Research Committee and Ethics Committee. The samples included in the study are identifiable by a lab-generated number that cannot be traced back to patients. Hence, all patient information was kept private. Also written consent for sample collection is asked from the patients and depending on the willingness of the patient; included in the study.
Study Area and Duration
This study was conducted in a single setting to assess the efficiency of EFDS considering EDVTM as the reference method. A total of 500 samples from Seven Hills Hospital, received for normal SARS CoV-2 testing, were used in the study. The dry swab was also collected at the same time from enrolled patients. After routine processing and reporting; dry swabs were processed with blinded ID as per protocol. The duration of the study was from June to July 2021.
Statistical Analysis
Statistical analysis guidelines followed as per Standards for Reporting Diagnostic Accuracy (STARD) recommendation. We also verified positive and negative agreement, especially for the qualitative findings. Also, differences between targets CT of the two methodologies were represented in a Bland-Altman plot by using MedCalc® statistical software version 20.009.
Methods
VTM-based Standard Protocol
Covid-19 RNA Extraction, PCR assay setup, and Result Interpretation: Nucleic acid extraction (200 uL sample input volume) was performed using MagMAX™ Viral/ Pathogen Nucleic Acid Isolation kit (Applied Biosystem -Thermo fisher) by using semi-automated Thermo Kingfisher Flex extraction system. PCR assay setup was done using QIAgility a liquid dispenser automated system by QIAgen; it mixes the RNA and master mix. COVIPATH RT-PCR kit and Quantstudio-5 (Thermo) was used for Amplification. The results were analyzed using Quantstudio design and analysis software version 1.5.1. All the procedures were done as per manufacture kit instruction.
Dry Swab Method
This method involves the processing of dry swabs from patients without extraction as per ICMR advisory. The sample is incubated for 30 minutes at room temperature with 400 μL of Tris-EDTA-Proteinase K buffer. About 50 μL of the sample is subjected to heat inactivation at 98ºc for 6 minutes. This extract is used for RT-PCR. The RNA and master mix were combined with the PCR assay setup using QIAgility a liquid dispenser automated system from QIAgen. COVIPATH RT-PCR kit and Quantstudio-5 (Thermo) were used for amplification. The results were analyzed using the Quantstudio application. All the procedures were done as per the manufacturer kit instruction.4
RESULTS
The flow and results of a performance characteristic of EFDS as compared to the reference method, EDVTM is as shown in Flowcharts 1 and 2.
Flowchart 1: Study design for evaluation of EFDS for detection of SARS CoV-2
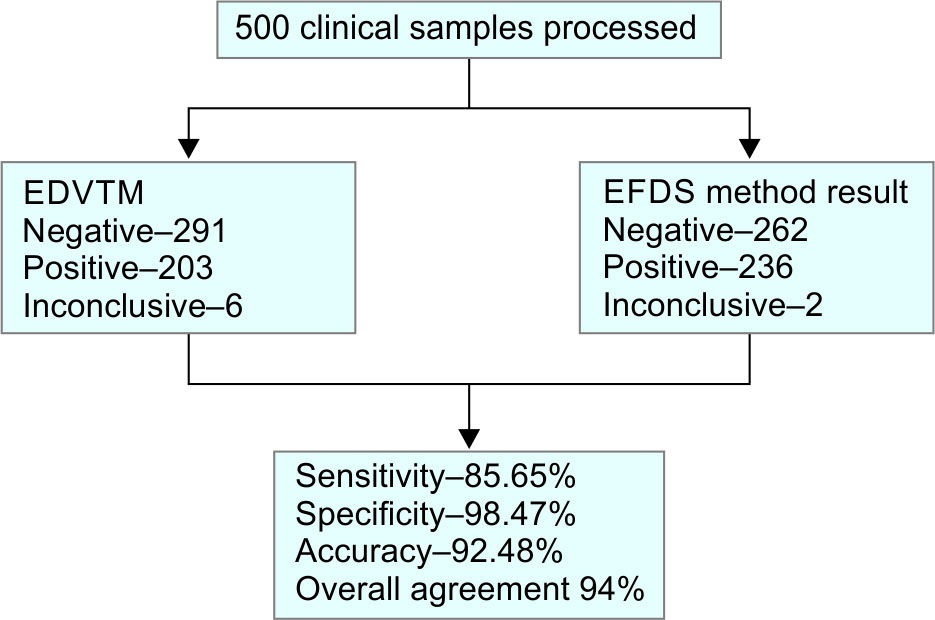
Flowchart 2: Enlistment and outcome of the study CoV-2
The overall performance of, EFDS as compared with EDVTM showed; 500 samples were used, and 455 of them produced concordant results, while 45 samples produced discrepant findings (Table 1). Upon analysis, the positive and negative agreements were estimated as 85.65% (CI: 80.44 to 89.91%) and 98.47% (CI: 96.14 to 99.58%) respectively (Table 2). Among 500 samples, SARS CoV-2 was detected in 197 samples (39%) and not detected in 258 (51%) by both the assays; There were 33(6.6%) samples not detected by EDVTM but detected by EFDS method and 4 (0.8%) samples not detected by the EFDS method and detected by EDVTM, 8 samples showed inconclusive result that is only one gene positive (after repeating) of which 6 samples were positive for both genes in EFDS Method and 2 Samples were positive for both genes in EDVTM Method.
| EFDS | |||||
|---|---|---|---|---|---|
| Positive | Negative | Inconclusive | Total | ||
| EDVTM | Positive | 197 | 4 | 2 | 203 |
| Negative | 33 | 258 | 0 | 291 | |
| Inconclusive | 6 | 0 | 0 | 6 | |
| Total | 236 | 262 | 2 | 500 | |
| Comparison | |
|---|---|
| Positive agreement | 85.65 (80.44–89.91) |
| Negative agreement | 98.47 (96.14–99.58) |
| Overall agreement | 92.48 (89.78–94.65) |
Bland-Altman Plot
This plot was drawn to analyze the extent of agreement between the assays (Fig. 1), and the heterogeneity between the CT values of ORF1ab gene and N gene of the two methods was plotted. Table 3 shows the mean difference between EFDS method and EDVTM for ORF1ab gene and N gene is –3.34 and –3.43 CT. The lower limit of agreement (LOA) for ORF1ab gene and N gene is –27.21 and –28.40 CT and the upper limit of agreement (ULA) for ORF1ab gene and N gene is 20.52 and 21.53 CT.
| Description | ORF1ab Gene | N Gene |
|---|---|---|
| Mean CT difference | –3.34 | –3.43 |
| Lower limit of agreement | –27.21 | –28.40 |
| Upper limit of agreement | 20.52 | 21.53 |
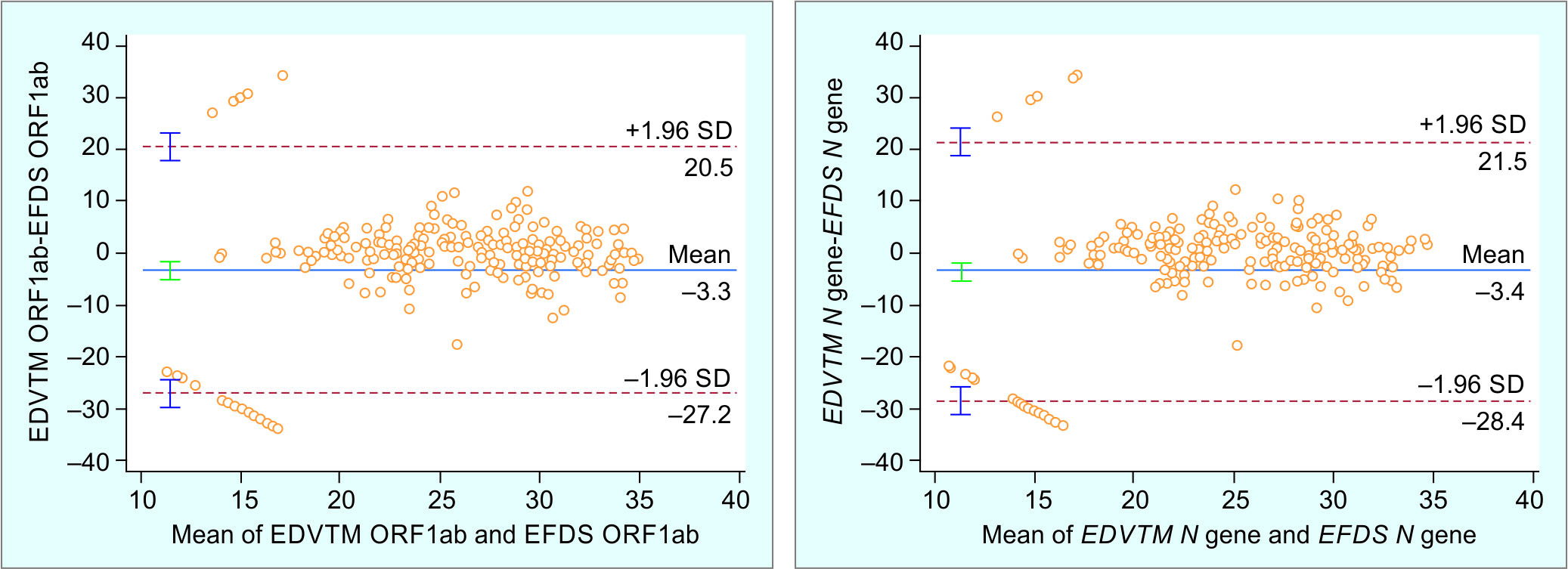
Fig. 1: Bland-Altman plot of targets ORF1ab and N-gene showing differences in EDVTM and EFDS
DISCUSSION
In order to control the spread of the infection, accurate identification of the infected individuals is the need of the hour. Given these various test results and the rapid spread of the infection, it is important that PCR should be widely practiced worldwide. Due to the ongoing pandemic, a massive number of swabs and PCR kits are required worldwide, placing a strain on resources and supply chains.8
The supply chains got significantly interrupted in the initial period of the pandemic itself, leaving laboratories struggling for any available commercial reserves. Several laboratories were driven to utilize different collection devices and transport media due to the pressure to enhance testing capacity, usually without conducting a standard test. Several scientists designed their own transport media because there weren’t any commercially accessible options.9 The existing protocol which was previously demonstrated to be acceptable was chosen because of the broad variety of specimens that can be transported.10
Collection and transportation of laboratory samples are executed with the use of alternative transport media.11 These media are not typically thought of as being the best option for the preservation of samples. However, owing to the shortage of viral and universal transport media (UTM), there was widespread use of alternative such media, which may not be ideal for the detection of the causative virus.
Recently Indian Council of Medical Research (ICMR) issued an advisory, for the detection of the causative virus by real-time reverse-transcription PCR, the overall sensitivity of this study 85.65% is more than that of the ICMR advisory for dry swabs which is 79% sensitivity and there is a slight decrease in the specificity 98.47% as compared to advisory 99%.4
In our study, variability was reported between the EFDS method and the reference method for 45 discordant results. As per prior research, a variety of possible reasons could cause assay discrepancies.12 Swabs collected in VTM are diluted with a VTM of 2 mL and the dry swabs are diluted with only 400 ul of buffers. There is five times more dilution of the viral load in VTM as compared with the EFDS method. Also, There are a variety of appropriate VTM formulations that might work for each laboratory’s specific set of conditions. While selecting a VTM formulation, the unique requirements of the shipping and receiving laboratories should be taken into account.
CONCLUSION
The EFDS assay showed a good correlation with the reference method. There was a slight increase in the detection rate showed by the EFDS assay as compared with EDVTM. As per these results; it can be concluded that EFDS method showed comparable performance with the comparative method and can be useful in for detection of SARS-CoV-2, EFDS method also reduces the cost and simplifies the workflow.
ACKNOWLEDGMENTS
Authors extend their regards to the colleagues who participated in this study. The study was sponsored by the Thyrocare Technologies Limited Team and Seven Hills Hospital.
ORCID
Prasad Tukaram Dhikale https://orcid.org/0000-0002-1860-6153
REFERENCES
1. National Administration of Traditional Chinese Medicine. The national health commission of the People’s Republic of China: corona virus disease 2019 diagnosis and treatment plan (5th trial edition revised version) [cited 2023 Mar 29]. Available from: http://english.www.gov.cn/.
2. Sharma K, Aggarwala P, Gandhi D, Mathias A, Singh P, Sharma S, et al. Comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: Quest for the best choice. PLoS One. 2021 Apr 5;16(4):e0249408. doi: 10.1371/journal.pone.0249408.
3. Qian Y, Zeng T, Wangh H. Safety management of nasopharyngeal specimen collection from suspected cases of coronavirus disease 2019 Int J Nurs Sci 2020;7:153–156. DOI: 10.3761/j.issn.0254e1769.2020.03.
4. Indian Council of Medical Research. Advisory on use of Dry Swab RNA Extraction Free RTPCR Method. [cited 2023 Mar 29]. Available from: https://www.icmr.gov.in/pdf/covid/techdoc/archive/Advisory_Dry_Swab_RNAExtraction_26112020.pdf.
5. Vermeiren C, Marchand-Senécal X, Sheldrake E, Bulir D, Smieja M, Chong S, et al. Comparison of Copan ESwab and FLOQSwab for COVID-19 diagnosis: working around a supply shortage. J Clin Microbiol 2020;58(6):e0.0669–e00620. DOI: https://doi.org/10.1128/JCM.00669-20.
6. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045.
7. Uhteg K, Jarrett J, Richards M, Howard C, Morehead E, Geahr M, et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. Journal of Clinical Virology. 2020 Jun 1;127:104384.CoviPath™ COVID-19 RT-PCR Kit USER GUIDE Multiplex real-time RT-PCR test intended for the qualitative detection of nucleic acid from SARS-CoV-2.
8. Nairz M, Bellmann-Weiler R, Ladstätter M, Schüllner F, Zimmermann M, Koller AM et al. Overcoming limitations in the availability of swabs systems used for SARS-CoV-2 laboratory diagnostics. Sci Rep 2021;11(1):2261. DOI: 10.1038/s41598-021-81782-8.
9. Centers for Medicaid and Medicare Services (CMS) CLIA Regulations and Federal Register Documents homepage. [cited 2023 Mar 29]. Available from: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/CLIA_Regulations_and_Federal_Register_Documents.
10. Centers for Disease Control and Prevention (CDC) SOP# DSR-052-01. Preparation of Viral Transport Medium homepage. [cited 2023 Mar 29]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf.
11. Indian Council of Medical Research. STANDARD PROTOCOL FOR BATCH TESTING OF OF VIRAL TRANSPORT MEDIUM (VTM) AND SWABS [cited 2023 Mar 29]. Available from: https://www.rgcb.res.in/documents/corona_alert/SOP-Batch%20Testing%20VTM.pdf.
12. Sakanashi D, Asai N, Nakamura A, Miyazaki N, Kawamoto Y, Ohno T, et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. Journal of Infection and Chemotherapy. 2021 Jan 1; 27(1):126–129. DOI: 10.1016/j.jiac.2020.09.027.
________________________
© The Author(s). 2023 Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and non-commercial reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.